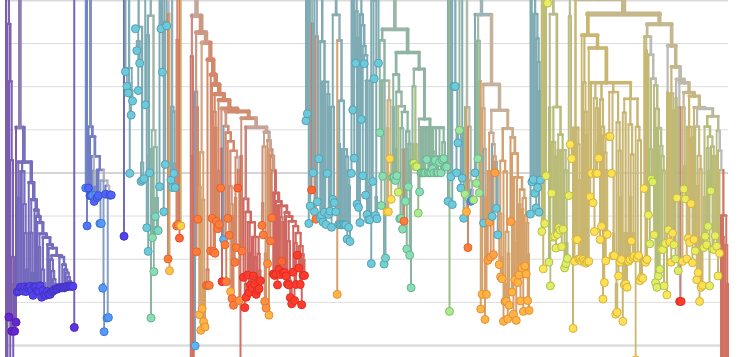
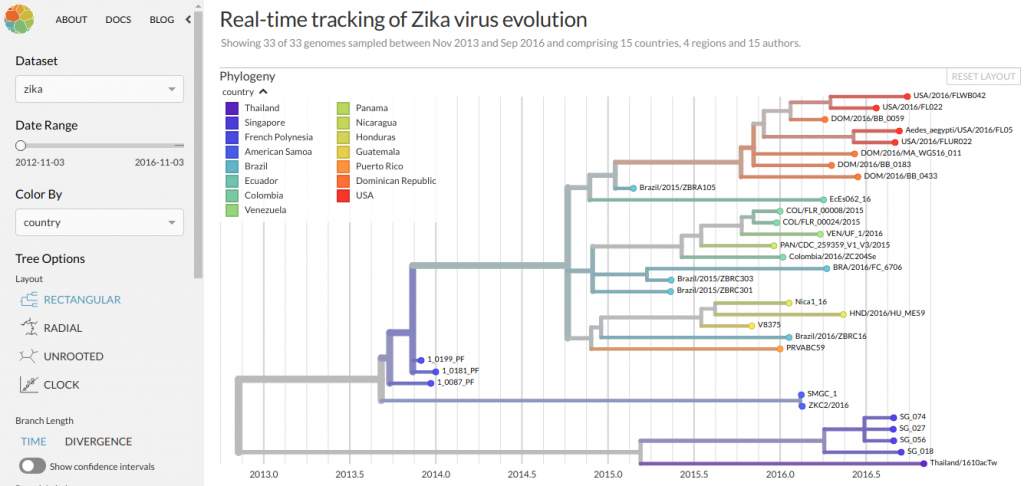
Nextstrain bioinformatics visualisation tool on Ubuntu server.
Nextstrain CLI – https://github.com/nextstrain/cli
Zika tutorial – Nextstrain Zika / Zika Tutorial / Getting started
Server installation
- Ubuntu server 18.04LTS
- on QEMU/KVM
- at least 4 CPU Cores
- Virtual network ‘default’ : NAT
Start and SSH to the Ubuntu Server
ssh user@192.168.122.156
Add universe repository
sudo add-apt-repository universe
Update system
sudo apt update sudo apt upgrade
Install Docker
sudo apt install apt-transport-https ca-certificates curl software-properties-common
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add -
sudo add-apt-repository "deb [arch=amd64] https://download.docker.com/linux/ubuntu bionic stable"
sudo apt update
apt-cache policy docker-ce
sudo apt install docker-ce
Docker without sudo
sudo usermod -aG docker ${USER}
su - ${USER}
id -nG
Check Python version
python3 --version
Install python3 PIP
sudo apt install python3-pip
Install nextstrain-cli
pip3 install nextstrain-cli
Reboot or logout/login
Check Nextstrain setup has been installed
nextstrain check-setup
Nextstrain Output
A new version of nextstrain-cli, 1.5.0, is available! You're running 1.4.1.
Upgrade by running:
pip install --upgrade nextstrain-cli
Testing your setup…
✔ docker is installed
✔ docker run works
All good!
Nextstrain commands
usage: nextstrain [-h] {build,view,deploy,shell,update,check-setup,version} ...
Nextstrain command-line tool
optional arguments:
-h, --help show this help message and exit
commands:
{build,view,deploy,shell,update,check-setup,version}
build Run pathogen build
view View pathogen build
deploy Deploy pathogen build
shell Start a new shell in the build environment
update Updates your local image copy
check-setup Tests your local setup
version Show version information
Install Zika data tutorial
Clone zika setup
git clone https://github.com/nextstrain/zika.git
Change to zika directory
cd zika
Build zika
nextstrain build .
On/If error copy example_data/zika.fasta to data folder
cp example_data/zika.fasta data/
Build zika again
nextstrain build .
View in auspice (Front-end)
nextstrain view auspice/ --allow-remote-access
Auspice output
——————————————————————————————————————————————————————————————————————————————
The following datasets should be available in a moment:
• http://192.168.122.156:4000/local/zika
——————————————————————————————————————————————————————————————————————————————
> auspice@1.29.0 server /nextstrain/auspice
> node server.js
-----------------------------------
Auspice server started on port 4000
Serving compiled bundle from /dist
Local datasets sourced from /nextstrain/auspice/data can be accessed via "/local/..." URLs
Local narratives sourced from /nextstrain/auspice/local_narratives can be accessed via "/local/narratives/..." URLs
-----------------------------------
Successfully created local manifest
Successfully created live manifest
Successfully created staging manifest
Charon API request: /charon?request=mainJSON&url=/local/zika
Success fetching v1 JSONs. Sending as a single JSON.
View datasets in the browser
http://192.168.122.156:4000/local/zika
Auspice start commands
usage: nextstrain view [-h] [--allow-remote-access] [--image <name>]
[--exec <prog>] [--auspice <dir>] [--docker-arg ...]
<directory>
Visualizes a completed pathogen build in auspice, the Nextstrain web frontend.
The data directory should contain sets of files with at least two files:
<prefix>_tree.json
<prefix>_meta.json
The viewer runs inside a container, which requires Docker. Run `nextstrain
check-setup` to check if Docker is installed and works.
positional arguments:
<directory> Path to pathogen build data directory
optional arguments:
-h, --help show this help message and exit
--allow-remote-access
Allow other computers on the network to access the
website (default: False)
development options:
These should generally be unnecessary unless you're developing build images.
--image <name> Container image in which to run the pathogen build
(default: nextstrain/base)
--exec <prog> Program to exec inside the build container (default:
auspice)
--auspice <dir> Replace the image's copy of auspice with a local copy
(default: None)
--docker-arg ... Additional arguments to pass to `docker run` (default:
None)